

During the COVID-19 pandemic, which generates a large proportion of asymptomatic cases, sewage monitoring can be a very valuable tool. The LIST, quickly set up a complete workflow, from sample collection to RT-qPCR detection (Luxembourg Institute of Health (LIH) protocols), to measure the circulation levels of SARS-CoV-2 in wastewater. Dr. Ogorzaly, who is leading the project, emphasises that “this impressive collaborative work would not have been possible without the valuable assistance and technical support of the Administration de la Gestion de l’Eau (AGE), the Ministry of Health, the Direction de la Santé, Aluseau (Association luxembourgeoise des services d’eau) and the different wastewater syndicates and municipalities of the country”.
By bringing together the four main microbiological research institutes of the country (Luxembourg Institute of Science and Technology (LIST), Laboratoire national de santé (LNS), Luxembourg Institute of Health (LIH), and the University of Luxembourg (uni.lu)), the CORONASTEP pilot project (FNR COVID-19 project, André Losch Fondation) proved wastewater monitoring to be an early, robust, and cost-effective detection system to support the Luxembourg authorities with reliable data. Thus, by sampling only 13 sites, the project monitored 73% of the population of Luxembourg (https://www.list.lu/en/covid-19/coronastep/). In addition, it has also demonstrated that it is possible to detect variants of concern while sequencing wastewater samples, as published in the journal Water (Herold et al 2021).
Indeed, new variants of interest (VOI) and of concern (VOC) of the SARS-CoV-2 appear regularly (Alpha, Beta, Gamma, Delta, Omicron…) and spread rapidly. They are characterised by specific combinations of mutations conferring increased transmissibility, virulence, or immune escape, which are threats to our response and efforts against this virus.
A mix of numerous variants
« Unlike the patient samples routinely analysed at the LNS, the wastewater samples have the particularity of being a mix of numerous variants whose genome may be degraded. This is the challenge of the CORONAVAR project, which requires numerous technical optimisations,” explains Dr. Fournier, coordinator of the project on the LNS side. Therefore, to address the complexity of wastewater as an analytical matrix, different complementary methodological approaches will be combined with real-time RT-PCR, namely the high throughput sequencing and the Digital Droplet RT-PCR (RT-ddPCR).
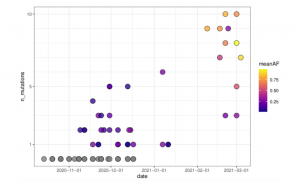
To examine wastewater samples and detect the presence of SARS-CoV-2 variants of concern, the LIST team has adapted the Digital Droplet RT-PCR (RT-ddPCR) technology for this purpose. By splitting the sample and reagents into a large number of droplets, this technology allows RT-PCR reactions to be performed on a single RNA molecule in each droplet. This permits the detection of rare mutations and improves the sensitivity of VOC detection 5- to 10-fold as well as the accuracy of quantification of SARS-CoV-2 variants in wastewater samples.
Previously tested in the CORONASTEP study, the next-generation amplicon sequencing of SARS-CoV-2 from wastewater samples can be done either on long fragments using Oxford Nanopore technologies (processed by the Luxembourg Centre for Systems Biomedicine (LCSB)) or on short fragments using Illumina sequencing (done by LNS). Both approaches have already resulted in a consensus sequence as well as an analysis of low-frequency variants (Herold et al 2021). However, “the bioinformatics processing of the sequencing data is more complex than for clinical samples, requiring optimisations from LNS’ Virology Sequencing team in order to increase the sequencing depth or set up the specific target enrichment”, explains Dr. Sibel Berger, responsible for the viral sequencing at LNS.
 |
|
The data collected during the CORONAVAR project will provide the prevalence of SARS-CoV-2 VOCs and VOIs in wastewater samples from approximately 75% of the Luxembourgish population. This original information is essential to help policy makers to manage the health crisis and to set up or lift different protective measures for the population based on the prevalence estimates at the wastewater treatment plants. In addition to the immediate support to the government, the project will contribute to the development of new tools and knowledge through the publication of scientific results and participation in international conferences.



