

On 24th November 2021, South Africa notified the World Health Organization (WHO) of the presence of a new SARS-CoV-2 lineage : B.1.1.529.
After a rapid assessment of the situation, the WHO designated it as a new variant of concern (VOC): Omicron*. 1
As of 29th November 2021 at 21:00 CET, 179 cases have been reported to the GISAID platform. 2 The first sample available so far was collected on 9th November in Gauteng, South Africa. This finding is accompanied by a rise in the number of cases reported in the country 3 (124 Omicron cases so far, representing more than 70% of the variants detected in November). 2
Other countries and regions that reported Omicron-positive cases are Botswana (19), Australia (5), Hong Kong (5), Canada (2) and Israel (1).
In Europe, specifically, 23 cases have been detected so far : 12 in the Netherlands (North-Holland), 4 in the United Kingdom (England), 4 in Italy (Campania and Lombardy), 1 in Belgium (Flemish Brabant), 1 in Germany (Hesse) and 1 in Austria (Tyrol) 2. No cases of the Omicron variant have been detected in Luxembourg so far. The geographical distribution of cases worldwide is shown in Figure 1.
The WHO classified the Omicron variant as VOC due to the high number of mutations found in its genome, especially in the region encoding the spike protein (up to 34 mutations, the most divergent so far)4. This protein is present on the surface of the virus and enables it to infect human cells. It is also the target of the main COVID-19 vaccines used worldwide. Therefore, changes in this protein might affect the behavior of the virus, especially its transmissibility, severity and vaccine effectiveness.
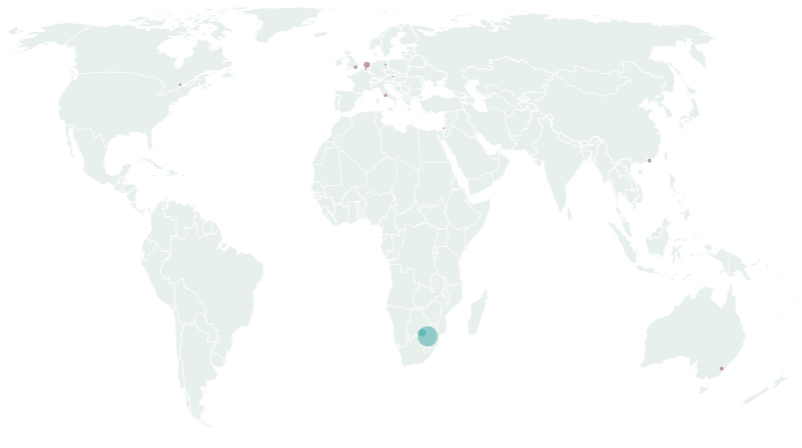
Figure 1 : Distribution of the Omicron cases notified to GISAID worldwide as of 29 Nov 2021 (map by Flourish®)
The information available so far is very limited. Hence, it is not yet sufficient to assess potential changes in transmissibility or severity, but preliminary data suggest a similar clinical behavior, with no unusual symptoms and including asymptomatic cases. 4,5
As for impact on vaccinated individuals, studies are still underway. 5 It is important to note that vaccines have proven to be effective against severe disease and death from previous variants, even if some vaccine breakthrough infections may occur. 3,5 However, given the uncertainty concerning the Omicron variant, to prevent new infections, it is of utmost importance now to comply with the well-known public measures of proven effectiveness: hand hygiene, face masks, cough/sneeze covering, physical distancing and ventilation. 6
The Omicron variant belongs to Pango lineage B.1.1.529, Nextstrain clade 21K and GISAID clade GR/484A. This variant is characterized by 45‐52 amino acid changes, including the aforementioned mutations to the spike protein, compared to the reference sequence. This could impact the sensibility of diagnostic tests, but the genome-based diagnostic tests already in use by laboratories in Luxembourg rely on the detection of multiple targets in the genome, not only the S gene, to prevent false negative results.
Interestingly, the Omicron variant has a diagnostic marker that was also present in the Alpha variant, which is the S-gene dropout or S-gene target failure (SGTF) due to a deletion at Spike position 69-70. This marker could be used as a rapid proxy test for this variant, pending sequencing confirmation, as there already exist commercial RT-PCR kits for its detection (e.g. Allplex® SARS-CoV-2 Variants I). Studies are ongoing to assess if the performance of Rapid diagnostic tests for detection of SARS‐CoV‐2 antigen may be impacted by this variant.
The LNS host the National Reference Laboratory for Acute Respiratory Infections, which runs the national SARS-CoV-2 genomic surveillance program to support Public Health actions through early detection and risk assessment. The specific objectives of this strategy are:
To do so, the LNS receives positive samples from hospitals and private laboratories and prioritises those from hospital cases, post-vaccination cases and clusters with high transmission. Finally, a systematic selection of all national samples is made to get a representative picture of the circulating variants.
The LNS also complies with the European Center for Disease Prevention and Control (ECDC) recommendations for a minimum proportion of samples to be sequenced (Figure 2). This enables the early detection of any emerging variant, with estimated anticipation of 7-8 weeks before it could potentially become the dominant variant. 7 Taking this into account, no cases of the Omicron variant have been detected during the month of November, but only Delta cases (including all the various AY subtypes).
Due to the emergence of the new Omicron variant, as well as the high incidence rates in the European context, the LNS is currently carrying a complementary sequencing effort to provide real-time monitoring of the circulating SARS-CoV-2 variants in the country. Besides the weekly reporting via the Respiratory Viruses Surveillance – ReViLux, 8 we will implement a flash sequencing and reporting for strategic samples: unusual transmission events (e.g. increased transmission despite the interventions in place), unexpected disease presentation/severity, vaccine-breakthrough infections and patients with known epidemiological link to settings where the Omicron variant is already circulating.
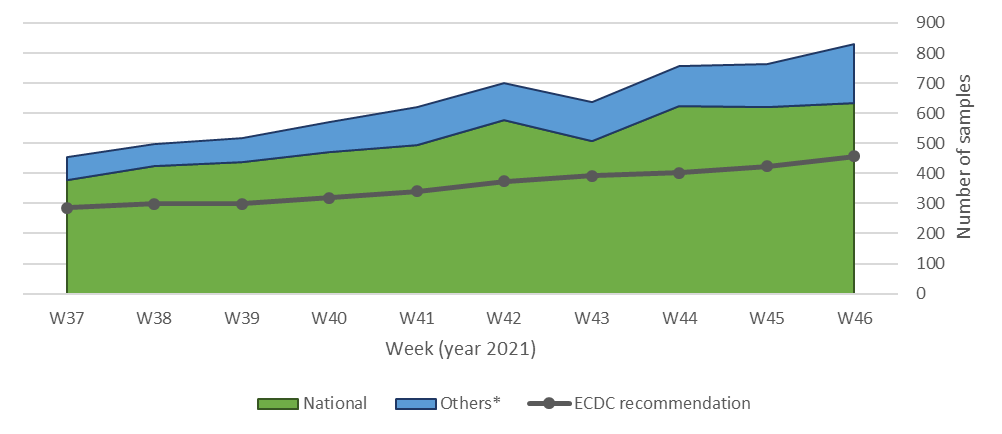
Figure 2. Number of samples sequenced in the LNS classified by the origin of the sample. *Others: including samples from non-residents and additional samples of interest.
Additionally, the genomic and molecular microbiology unit (LUX-GEMM) at LNS will introduce a large scale screening strategy based on the aforementioned SGTF approach to enable faster detection and identification of any potential Omicron variant cases. All positive samples referred to the LNS as National Reference Laboratory for Acute Respiratory Infection will be screened by RT-PCR in real-time (within 24 h of reception), and all potential cases will be confirmed by our sequencing platform.
Finally, the epidemic intelligence unit will continue its collaboration with neighboring countries and international public health organizations (e.g. ECDC and WHO) to ensure the timely reporting of genomic data to the public domain and inform any public health actions, so that appropriate clinical care can be maintained and contact tracing teams are timely informed to break chains of transmission.
* Two letters in the Greek alphabet were skipped, Nu (too similar to the English word “new”) and Xi (a common Chinese surname), following the WHO Best Practices for the Naming of New Human Infectious Diseases. 9