The Sentinel Surveillance Network identified 2 cases of influenza-like illness, thus remaining below the recommended threshold for the interepidemic season, according to the European Center for Disease Prevention and Control (ECDC) guidelines.
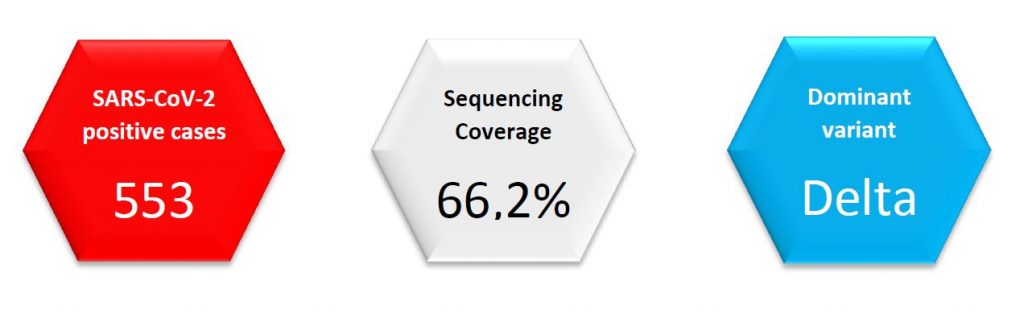
Regarding SARS-CoV-2 genomic surveillance, the Laboratoire national de santé analysed 366 specimens from residents in Luxembourg in week 36/2021 (from 553 total cases in the Grand Duchy of Luxembourg, 66,2%). This exceeds the minimum coverage (10%) and minimum sample size (288) recommended by the ECDC.
All specimens from week 36/2021 but one were assigned to the Delta variant, the remaining being Alpha variant. Community surveillance showed that parental lineage B.1.617.2 continues to be the most frequent one (77,1%), followed by AY.12 (11,1%). In respect to target groups, AY lineages were found in 25,8% of post-vaccination breakthrough cases.
The Laboratoire national de santé, as National Reference Laboratory for Acute Respiratory Infections in Luxembourg, performs close surveillance on respiratory viruses, with a special focus on SARS-CoV-2. There are currently three active projects:
The Sentinel Surveillance Network. It provides a broad picture of respiratory diseases affecting the Luxembourgish population, based on its double monitoring system (syndromic and virological).
The National SARS-COV-2 Genomic Surveillance Program. It enables detailed observation of SARS-CoV-2 mutations and variants through time and space, and also monitoring specific groups of interest.
The COVVAC Serology Project. It assesses the post-vaccination serological status in long-term care facilities and its evolution over time.
The ReViLux provides updates on the first two projects.
The Sentinel Surveillance Network aims at monitoring the circulating respiratory viruses, including SARS-CoV-2, and hence underpin public health actions. Following the World Health Organization (WHO) and European Centre for Disease Prevention and Control (ECDC) guidance, it focuses on cases of acute respiratory infection (ARI) and influenza-like illness (ILI).
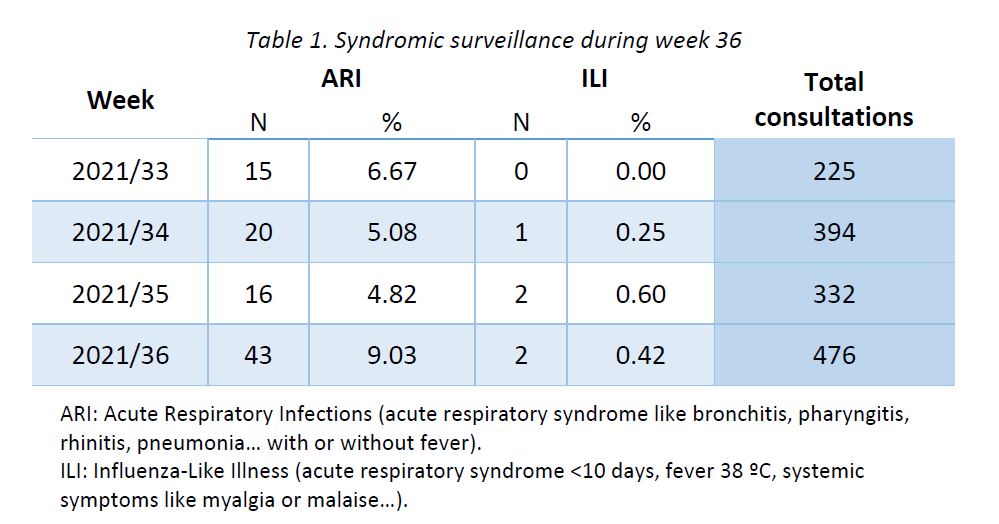
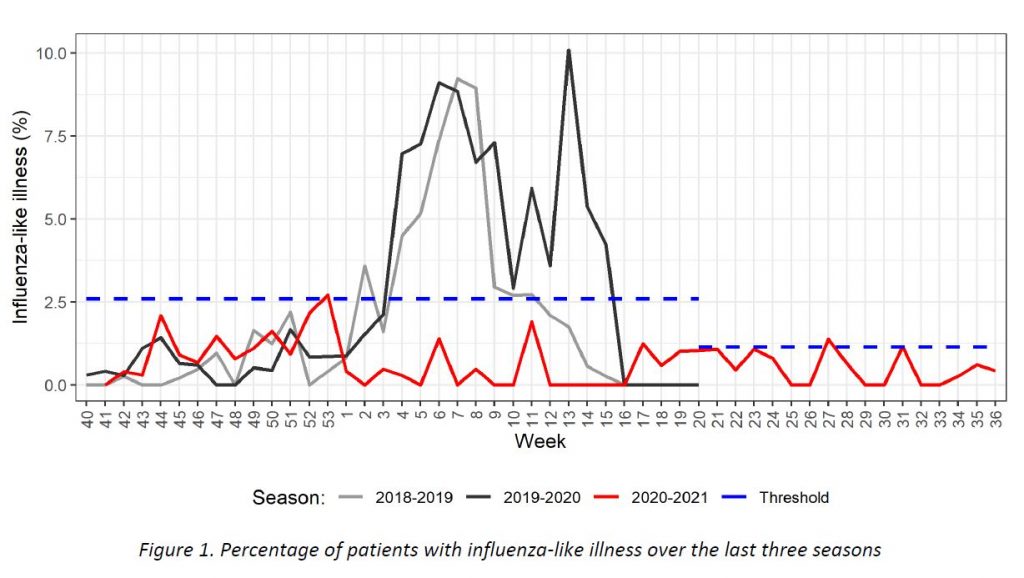
Results of syndromic surveillance during week 36 are displayed in Table 1 and the history of ILI consultations since the 2018-2019 season is shown in Figure 1. Two cases of ILI were identified in week 36 (out of 476 consultations); therefore, the percentage of ILI (0,42%) remains below the threshold for the interepidemic season (1,14%), according to the ECDC.
Regarding the virological surveillance, no data is available for week 36.
The National Reference Laboratory for Acute Respiratory Infections at LNS receives SARS-CoV-2 positive samples (nasopharyngeal or oropharyngeal swabs analysed by RT-PCR) from the national network of laboratories and proceeds as follows:
* Sequencing all specimens from hospital cases.
* Sequencing all specimens from reinfection and post-vaccination cases.
* Sequencing all specimens from cluster cases.
* Sequencing a representative sample of community cases.
The representative sample of community cases is a systematic selection from all SARS-CoV-2 positive cases registered in Luxembourg to detect emerging variants and early increases in their incidence and transmission within the community in Luxembourg. This sample is selected according to the ECDC guidelines.
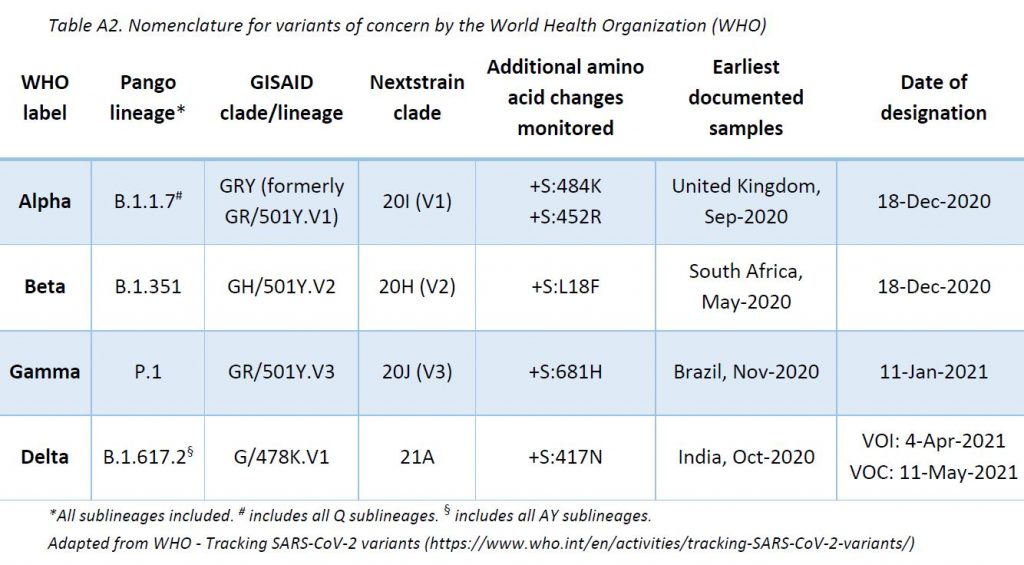
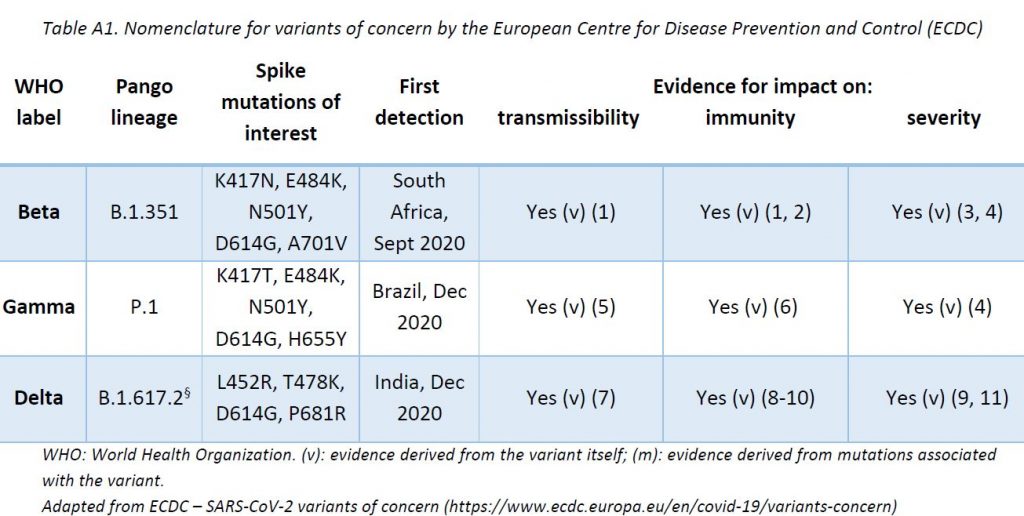
The LNS shares its sequencing results with GISAID EpiCov database (www.gisaid.org) periodically. SARS-CoV-2 lineages (variants) have been assigned based on Rambaut et al. using Phylogenetic Assignment of Named Global Outbreak LINeages (pangolin) software (v3.1.11, pangoLEARN 2021-08-24). The ReViLux continues to use the Pango nomenclature, in addition to the WHO nomenclature, to allow easier visualization of links between any evolving variants and their ancestor (https://cov-lineages.org). See nomenclature equivalences in Annex 1.
Methodological note on Delta subtypes
Since 3 September 2021, the ECDC no longer considers B.1.1.7 and B.1.1.7+E484K lineages (Alpha variant) as variants of concern. This decision is based in both their low circulation and the high effectiveness of vaccines in controlling them.
Since 24 August 2021, the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) software uses a new version of its reference library (pango-designation v1.2.66), which includes 13 additional Delta lineages (AY.13 to AY.25). Both B.1.617.2 and AY lineages are classified as Delta variant by the World Health Organization.
In week 36, 553 new cases were registered in Luxembourg; hence, the minimum sample size required to detect emerging variants at a 2.5% incidence is estimated to be 288 specimens (52.1%).
The microbial genomics unit at the LNS sequenced 475 specimens from week 36, with 366 specimens having been collected in week 36 from residents (66,2% coverage of the 553 total cases registered in Luxembourg; see coverage in Figure 2). This exceeds the minimum coverage (10%) and minimum sample size (288) to detect a 2.5% incidence recommended by the ECDC. The representative sample of community cases is built by systematic selection.
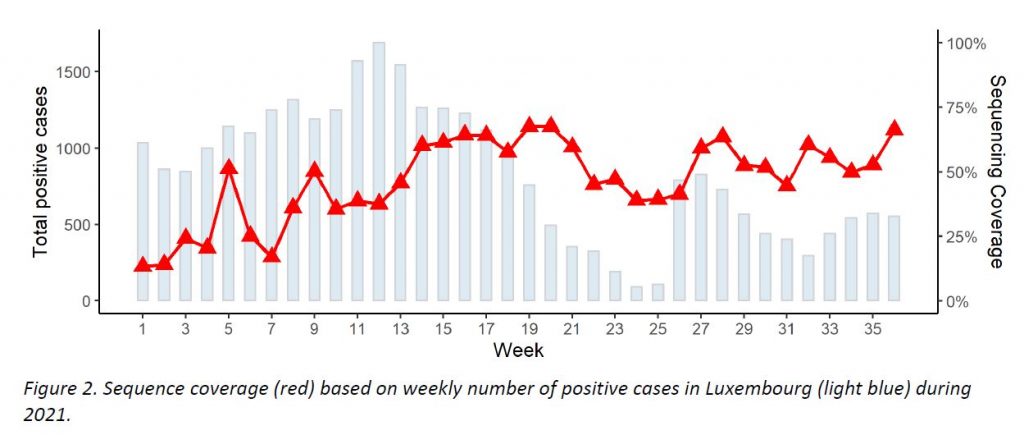
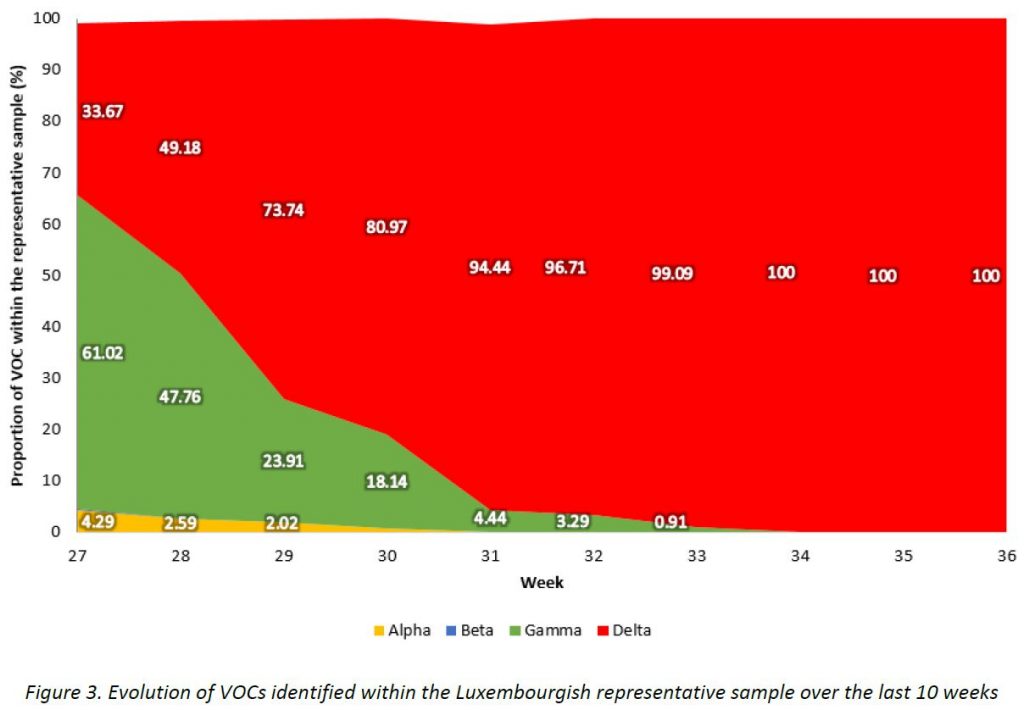
The evolution of variants over the weeks is shown in Figure 3.
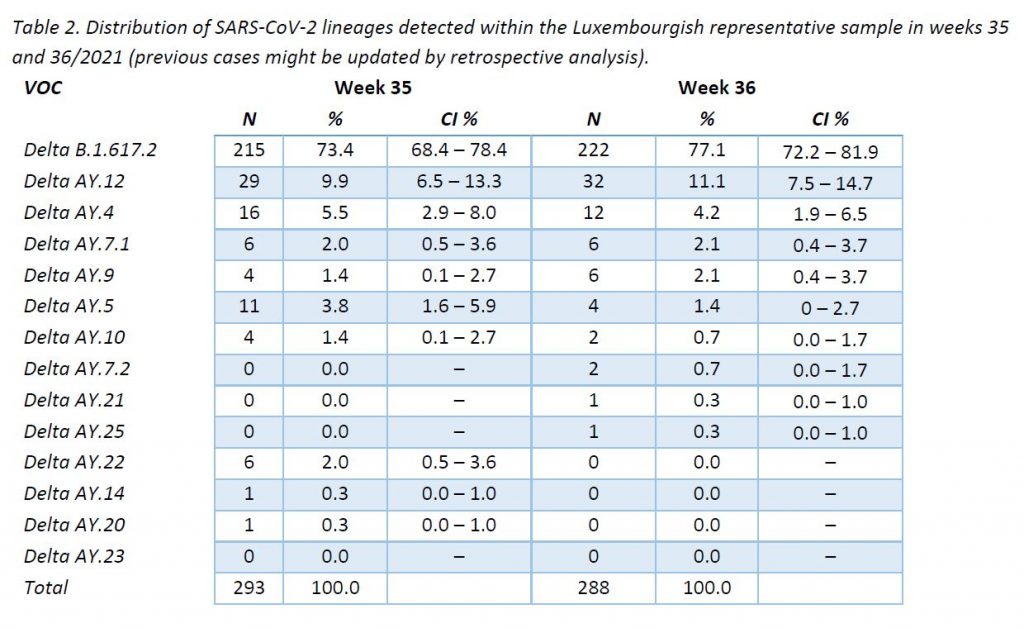
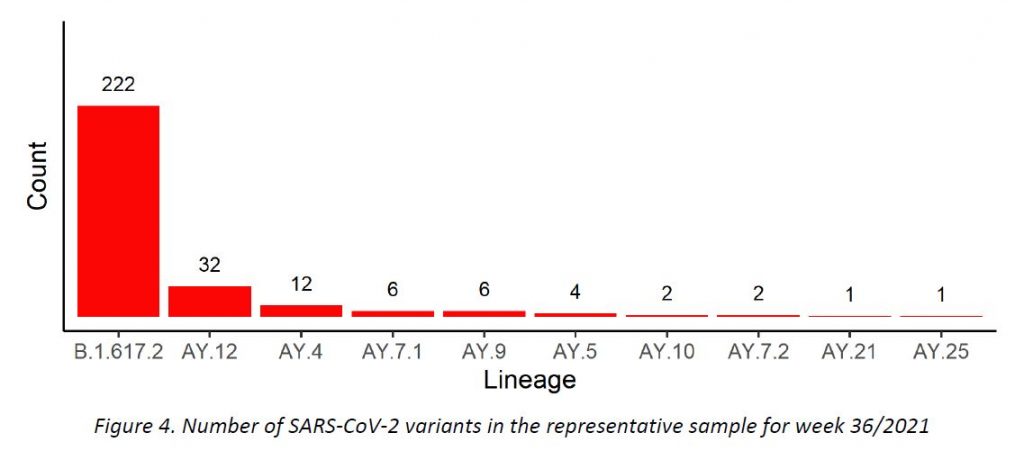
In week 36/2021, only Delta variant cases were detected among all specimens sequenced in the representative sample of community cases, including 9 Delta subtypes. The distribution of VOCs is displayed in Table 2, and their evolution over the weeks is shown in Figure 4.
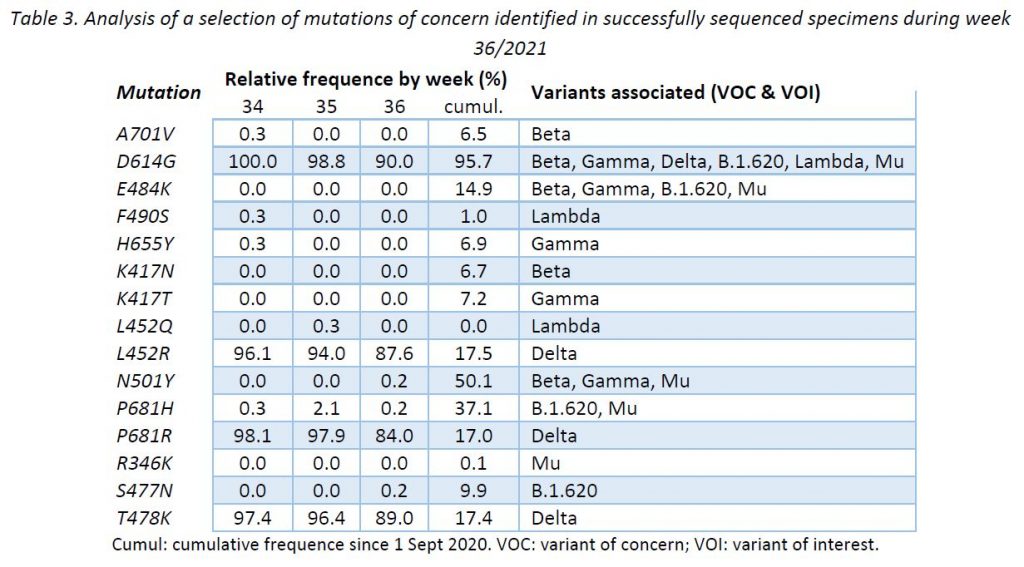
In addition to the surveillance of SARS-CoV-2 variants, the LNS monitors the occurrence of SARS-CoV-2 mutations reported to have a clinical and epidemiological relevance. This complementary surveillance enables us to detect unexpected mutations among the specimens sequenced. It is expected that VOC defining mutations share the same distribution as their corresponding VOCs. However, newly acquired mutations may occur and their early detection might be key to expect changes in the epidemic evolution.
Following ECDC guidance, the LNS is currently monitoring 15 mutations to the spike protein frequently associated to VOCs and variants of interest (VOIs). Table 3 provides the cumulative frequencies of these mutations, detected in the lineage-assignable genome sequences since 1 Sep 2020 (N = 18 231), as well as the frequencies over the last 3 weeks.
Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Retrieved 20 September 2021, from https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
COVID-19 Data Portal – accelerating scientific research through data. (2021). Retrieved 20 September 2021, from https://www.covid19dataportal.org/sequences
European Centre for Disease Prevention and Control. Guidance for representative and targeted genomic SARS-CoV-2 monitoring – 3 May 2021. ECDC : Stockholm ; 2021
European Centre for Disease Prevention and Control. SARS-CoV-2 variants of concern. Retrieved 20 September 2021, from https://www.ecdc.europa.eu/en/covid-19/variants-concern
Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. Geneva: World Health Organization; 2021.
GitHub – cov-lineages/pangolin: Software package for assigning SARS-CoV-2 genome sequences to global lineages. (2021). Retrieved 20 September 2021, from https://github.com/cov-lineages/pangolin
Hadfield J., Megill C., Bell S., Huddleston J., Potter B., Callender C. et al. (2018). Nextstrain: real-time tracking of pathogen evolution. Bioinformatics, 34(23), 4121-4123. doi: 10.1093/bioinformatics/bty407
Pango Network. New AY lineages. Retrieved 20 September 2021, from: https://www.pango.network/new-ay-lineages/
Pango Network. New AY lineages. Retrieved 20 September 2021, from: https://www.pango.network/new-ay-lineages-and-an-update-to-ay-4-ay-12/
Rambaut A., Holmes E., O’Toole Á., Hill V., McCrone J., Ruis C. et al. (2020). A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiology, 5(11), 1403-1407. doi: 10.1038/s41564-020-0770-5
References: