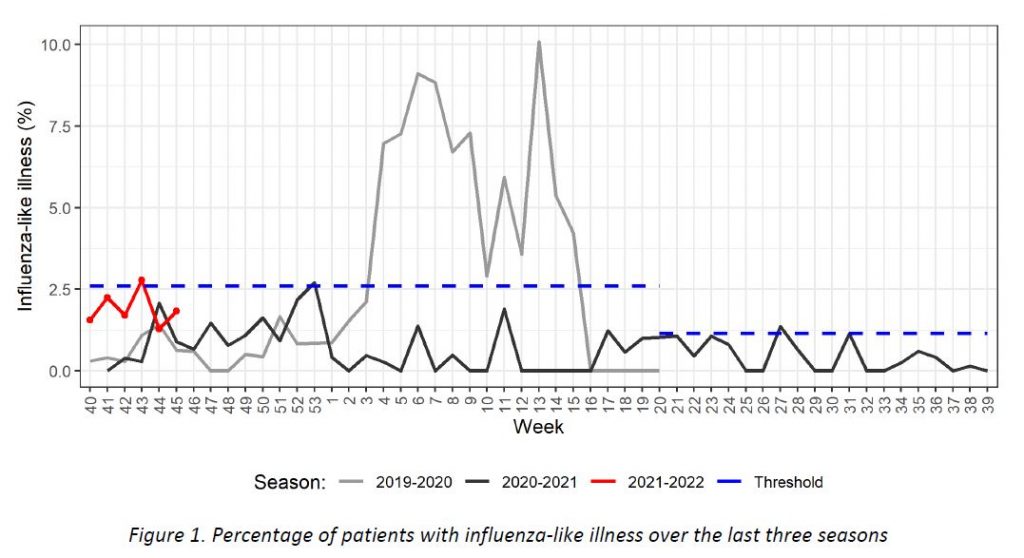
The Sentinel Surveillance Network identified 9 cases of influenza-like illness, thus remaining below the recommended threshold for the new epidemic season, according to the European Center for Disease Prevention and Control (ECDC) guidelines.
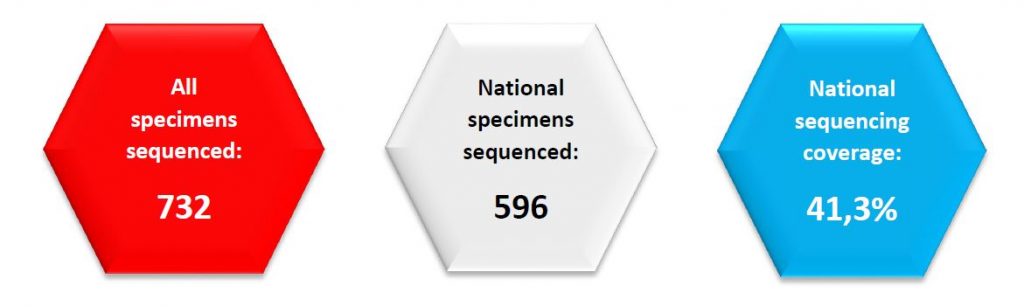
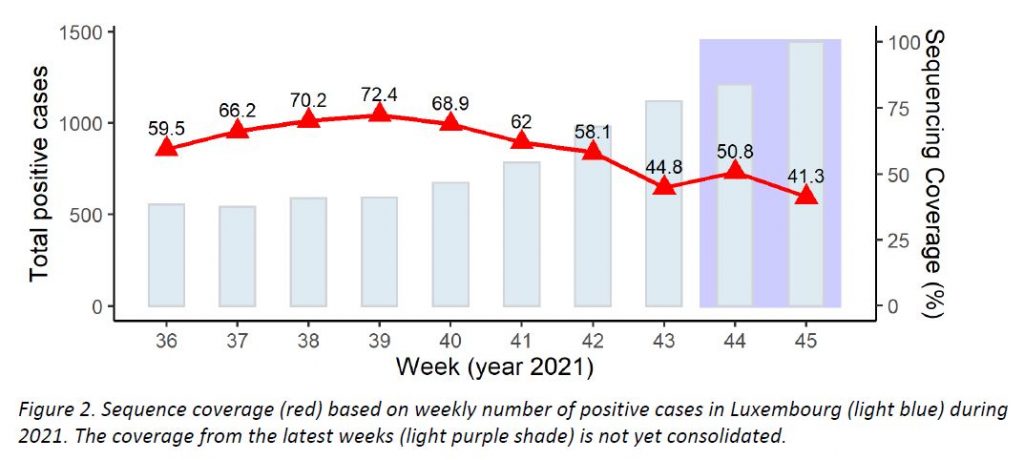
Regarding SARS-CoV-2 genomic surveillance, the Laboratoire national de santé analysed 596 specimens from residents in Luxembourg in week 45/2021 (from 1444 total cases in the Grand Duchy of Luxembourg, 41,3%). This exceeds the minimum sample size (424) recommended by the ECDC to detect emerging variants of a 2,5% proportion.
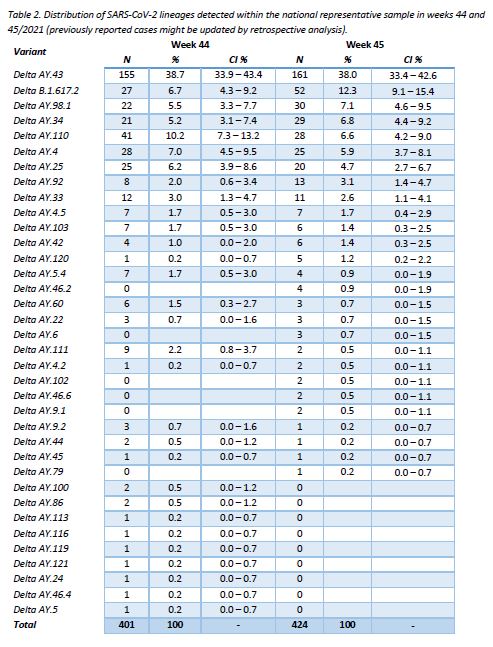
All specimens were assigned to the Delta variant. Community surveillance showed that the AY.43 lineage continues to be the most frequent one (38,0%), followed by B.1.617.2 (12,3%) and AY.98.1 (7,1%). Regarding target groups, the same three lineages were found to be the most frequent ones, both within hospital specimens and post-vaccination breakthrough cases. As for the mutations under surveillance, they revealed no outstanding behaviour, remaining in agreement with the lineages observed.
The Laboratoire national de santé, as National Reference Laboratory for Acute Respiratory Infections in Luxembourg, performs close surveillance on respiratory viruses, with a special focus on SARS-CoV-2. There are currently two active projects on which the ReViLux provides updates:
The Sentinel Surveillance Network. It provides a broad picture of respiratory diseases affecting the Luxembourgish population, based on its double monitoring system (syndromic and virological).
The National SARS-COV-2 Genomic Surveillance Program. It enables detailed observation of SARS-CoV-2 mutations and variants through time and space, and also monitoring specific groups of interest.
The Sentinel Surveillance Network aims at monitoring the circulating respiratory viruses, including SARS-CoV-2, and hence underpin public health actions. Following the World Health Organization (WHO) and European Centre for Disease Prevention and Control (ECDC) guidance, it focuses on cases of acute respiratory infection (ARI) and influenza-like illness (ILI).
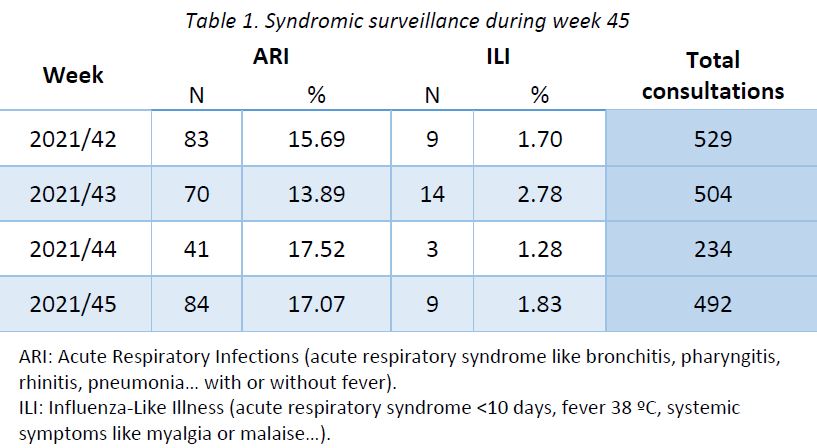
Week 40 marked the beginning of the new influenza season 2021-2022. Results of syndromic surveillance during week 44 are displayed in Table 1 and the history of ILI consultations since the 2019-2020 season is shown in Figure 1. Nine cases of ILI were identified in week 45 (out of 492 consultations); therefore, the percentage of ILI (1,83%) remains below the threshold for the epidemic season (2,59%), according to the ECDC.
Regarding the virological surveillance, a partnership among the CNS, private laboratories and the LNS recently started and will enable us to monitor the presence of several respiratory viruses. Results from the first analyses will be published soon.
The National Reference Laboratory for Acute Respiratory Infections at LNS receives SARS-CoV-2 positive samples (nasopharyngeal or oropharyngeal swabs analysed by RT-PCR) from the national network of laboratories and proceeds as follows:
Sequencing all specimens from hospital cases.
Sequencing all specimens from post-vaccination cases.
Sequencing specimens from clusters with high transmission.
Sequencing a representative sample of community cases.
The representative sample of community cases is a systematic selection from all SARS-CoV-2 positive cases registered in Luxembourg to detect emerging variants and early increases in their incidence and transmission within the community in Luxembourg. This sample is selected according to the ECDC guidelines.
The LNS shares its sequencing results with GISAID EpiCov database (www.gisaid.org) periodically. SARS-CoV-2 lineages (variants) have been assigned based on Rambaut et al. using the Phylogenetic Assignment of Named Global Outbreak LINeages (pangolin) software (v3.1.16, pangoLEARN 2021-11-09). Delta lineages nomenclature is under constant review. The original Delta B.1.617.2 lineage is being re-classified into more specific AY lineages in order to enable a more precise tracing of the cases. This report is based on the latest nomenclature, and previously assigned lineages have been updated to remain consistent with the latest nomenclature.
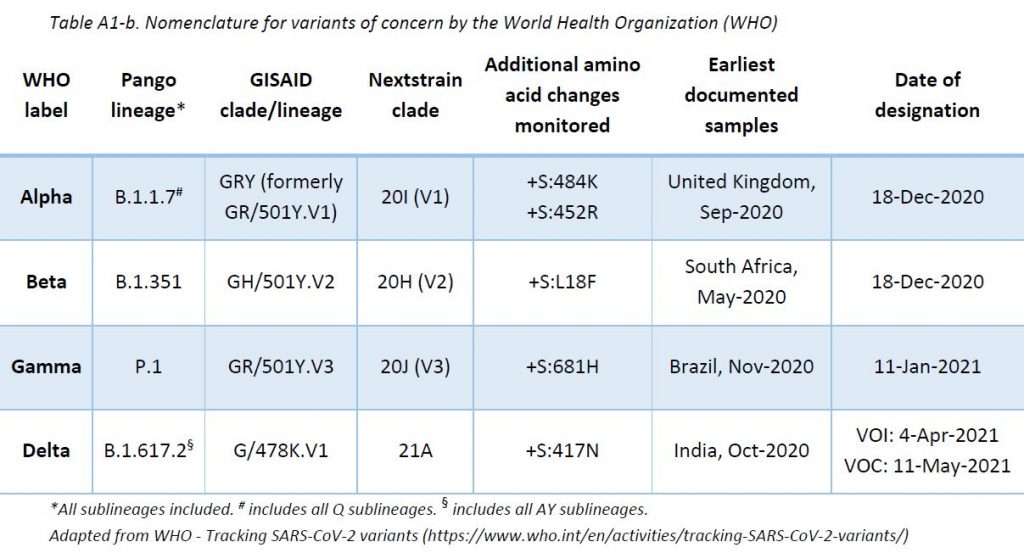
The ReViLux continues to use the Pango nomenclature, in addition to the WHO nomenclature, to allow easier visualization of links between any evolving variants and their ancestor (https://cov-lineages.org). See nomenclature equivalences in Appendix 1.
Methodological notes
Alpha variant is no longer considered as a variant of concern by the ECDC, due to both its low circulation and the high effectiveness of vaccines in controlling it. Therefore, specimens assigned to this variant are no longer reported separately.
In week 45, 1444 new cases were registered in Luxembourg; hence, the minimum sample size required to detect emerging variants at a 2,5% incidence is estimated to be 424 specimens (29,4%).
As shown in Figure 2, last week the microbial genomics unit at the LNS analysed 732 specimens from week 45, with 596 specimens having been collected in week 45 from residents (41,3% coverage of the 1444 total cases registered in Luxembourg; see coverage trend in Figure 2). This exceeds the minimum sample size (424) to detect a 2,5% incidence recommended by the ECDC. The representative sample of community cases is built by systematic selection.
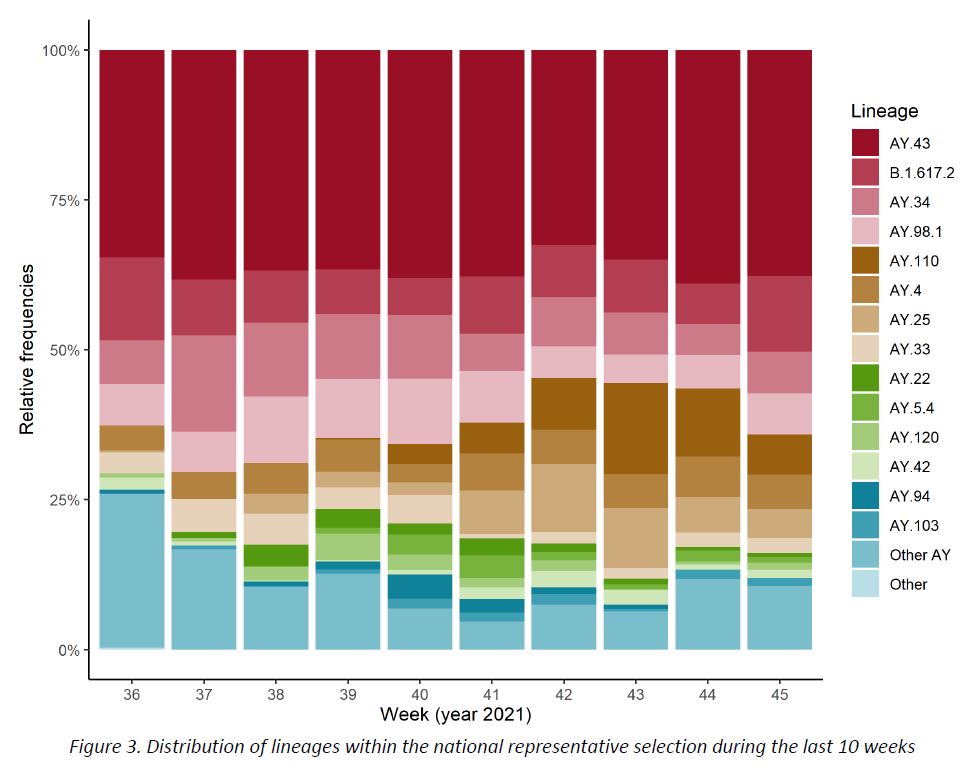
The distribution of successfully assigned lineages within the representative sample is shown in Figure 3. The selection of displayed lineages is based on their prevalence during the last 10 weeks (min. 1%). This distribution is further detailed for the last 2 weeks in Table 2. The lineage AY.43 continues to be the most frequent one (38,0%), followed by B.1.617.2 (12,3%), while the remaining lineages show frequencies lower than 10%.
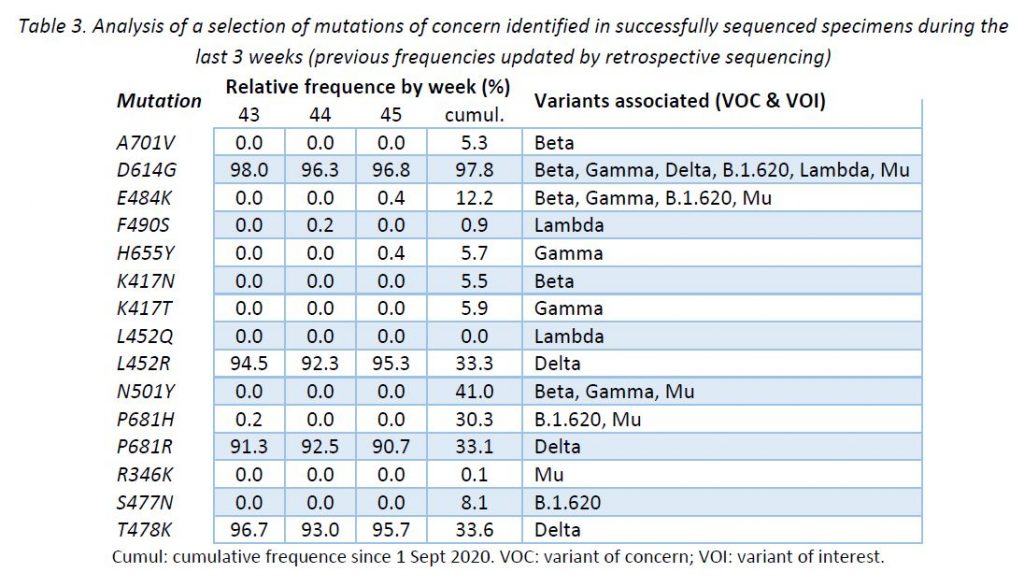
In addition to the surveillance of SARS-CoV-2 variants, the LNS monitors the occurrence of SARS-CoV-2 mutations reported to have a clinical and epidemiological relevance. This complementary surveillance enables us to detect unexpected mutations among the specimens sequenced. It is expected that VOC defining mutations share the same distribution as their corresponding VOCs. However, newly acquired mutations may occur and their early detection might be key to expect changes in the epidemic evolution.
Following ECDC guidance, the LNS is currently monitoring 15 mutations to the spike protein frequently associated to VOCs and variants of interest (VOIs). Table 3 provides the cumulative frequencies of these mutations, detected in the lineage-assignable genome sequences since 1 Sep 2020 (N = 21 506), as well as the frequencies for the last 3 weeks.
In broad terms, the mutations identified were expected according to the lineages assigned during this week. No mutation showed an outstanding behaviour.
Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Retrieved 22 November 2021, from https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
COVID-19 Data Portal – accelerating scientific research through data. (2021). Retrieved 22 November 2021, from https://www.covid19dataportal.org/sequences
European Centre for Disease Prevention and Control. Guidance for representative and targeted genomic SARS-CoV-2 monitoring – 3 May 2021. ECDC : Stockholm ; 2021
European Centre for Disease Prevention and Control. SARS-CoV-2 variants of concern. Retrieved 22 November 2021, from https://www.ecdc.europa.eu/en/covid-19/variants-concern
Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. Geneva: World Health Organization; 2021.
GitHub – cov-lineages/pangolin: Software package for assigning SARS-CoV-2 genome sequences to global lineages. (2021). Retrieved 22 November 2021, from https://github.com/cov-lineages/pangolin
Hadfield J., Megill C., Bell S., Huddleston J., Potter B., Callender C. et al. (2018). Nextstrain: real-time tracking of pathogen evolution. Bioinformatics, 34(23), 4121-4123. doi: 10.1093/bioinformatics/bty407
Pango Network. New AY lineages. Retrieved 22 November 2021, from: https://www.pango.network/new-ay-lineages/
Pango Network. New AY lineages. Retrieved 22 November 2021, from: https://www.pango.network/new-ay-lineages-and-an-update-to-ay-4-ay-12/
Rambaut A., Holmes E., O’Toole Á., Hill V., McCrone J., Ruis C. et al. (2020). A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiology, 5(11), 1403-1407. doi: 10.1038/s41564-020-0770-5
According to the ECDC
References:
According to the WHO