- The Laboratory
- Organization
- Departments
- Jobs
- Analysis book
- Contact
- News
- Publications
- Download


A new SARS-CoV-2 variant referred to as SARS-CoV-2 VUI 202012/01 (Variant Under Investigation, year 2020, month 12, variant 01), has been declared by the British authorities. The variant was detected via genomic sequencing in the United Kingdom (UK). It is defined by multiple spike protein mutations (deletion 69-70, deletion 144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) present.
The new variant is correlated with a rapid increase in COVID-19 incidence. This increase was reported in South East England. Analysis using viral genome sequence data identified a large proportion (>50%) of cases that belonged to a new single lineage. Overall, around 5 to 10% of all COVID-19 cases are regularly sequenced in the UK. As of 26 December 2020, more than 3000 individuals had been identified with this virus variant in England, with the earliest case identified from 20 September 2020. Several countries had reported sporadic cases caused by the new variant from other EU/EEA countries (Belgium, Denmark, France, Germany, Iceland, Ireland, Italy, the Netherlands, Norway, Portugal, Spain and Sweden) and globally (Australia, Canada, Hong Kong SAR, Israel, Japan, Jordan, Lebanon, South Korea, Switzerland, Singapore).
South Africa reported another SARS-CoV-2 variant of potential concern, designated 501.V2 that was first observed in samples from October and since then evolved to be the dominant circulating variant in South Africa.
In response to the emergence of the new variant several countries had issued a travel ban from the UK raising many concerns on the consequence of the transmission to this variant globally.
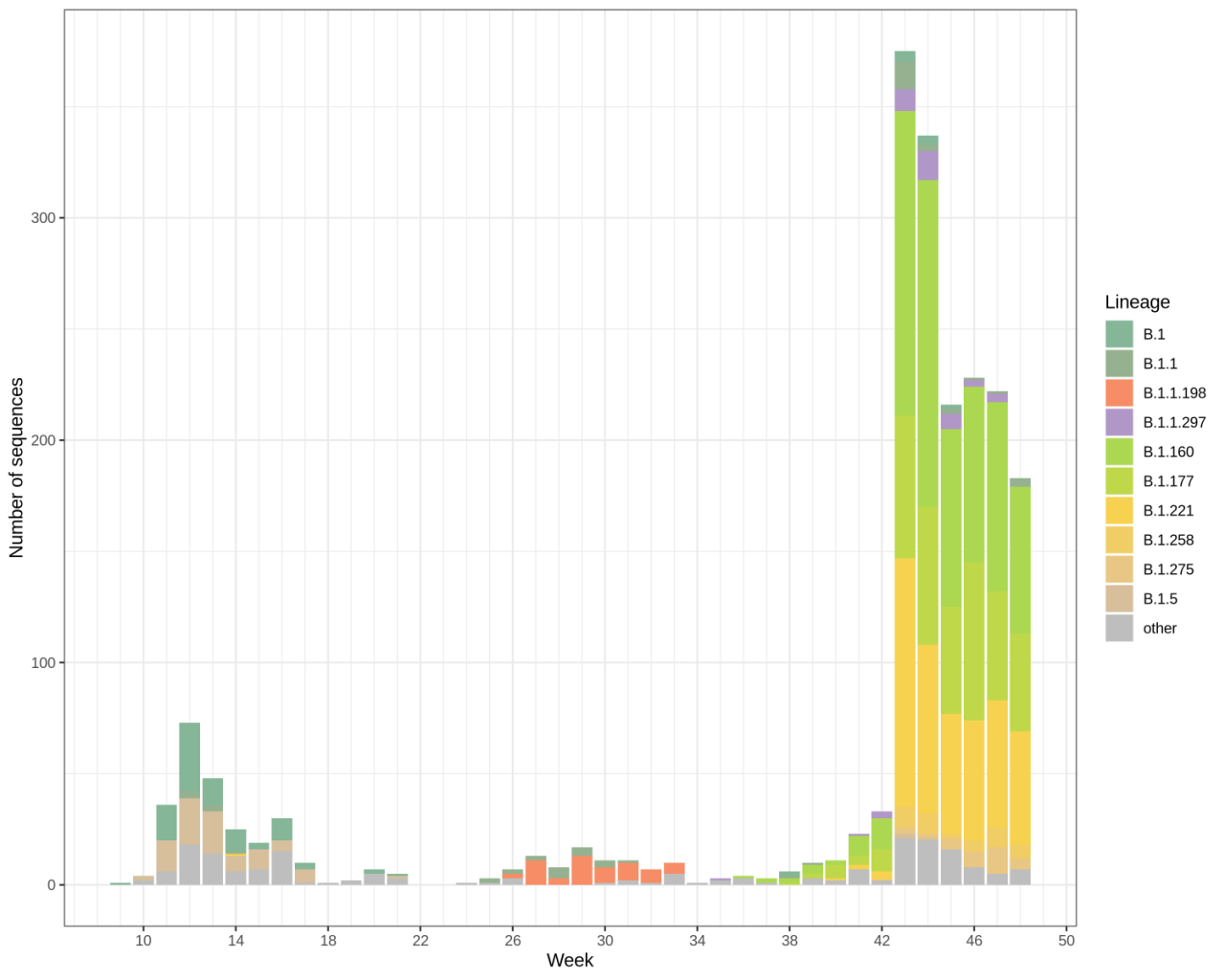
The microbiology department at the Laboratoire national de santé (LNS) sequenced 3027 SARS-CoV-2 positive samples since 29/02/2020, wherein 2285 have been added to the sequencing pool since 01/10/2020. We currently had increased our sequencing capacity to reach 10% of community circulating variants.
Most recent sequencing data referring to week 50 and 51 confirm observations made since the beginning of September of three main circulating strains. It is to note that annotation data have been updated, so that lineages have been added, might have been regrouped, or renamed. This mainly applies to the high-prevalent Dutch lineage B.1.78, which disappeared, but has been replaced by lineage B.1.221 (22%; Netherlands/Belgium). Accordingly, the geographical source stays consistent. The other two main circulating strains remain as observed in previous weeks; lineages B.1.160 (32%), B.1.177 (38%).
Lineage B.1.160 represents an EU/EEA and UK multi-country cluster which involves Belgium, France, Germany, and the United Kingdom. It is characterized by a spike protein mutation S: S477N, occurring in combination with N: M234I, A376T, ORF1a: M3087I, ORF1b: A176S, and most also with ORF1b: V767L K1141R, E1184D.
Lineage B.1.177 appeared in early summer 2020 and has its origin presumably in Spain. It spread to multiple European countries and its frequency rose to 40-70% in Switzerland, Ireland, and the United Kingdom in September. It is as well prevalent in other Luxembourg neighboring countries like Netherlands and France. It has the mutations A222V in the spike protein and A220V in the nucleoprotein.
The new UK variants attribute lineage B1.1.7 was not detected in our last analysis on samples collected until 18/12/2020. We are currently sequencing samples from week 51 to investigate the appearance of the variant in Luxembourg.
Since the emergence of the new variant and the declaration of the increased transmissibility (up to 70%), various implications for human health have been considered:
In Luxembourg, LNS will work closely with key stakeholders to implement the ECDC recently published recommendations as follows:
It is essential to reiterate the importance of strict adherence to the sanitary measures implemented in Luxembourg during the festive periods in particular guidance on the avoidance of non-essential travel and social activities.