- The Laboratory
- Organization
- Departments
- Jobs
- Analysis book
- Contact
- News
- Publications
- Download


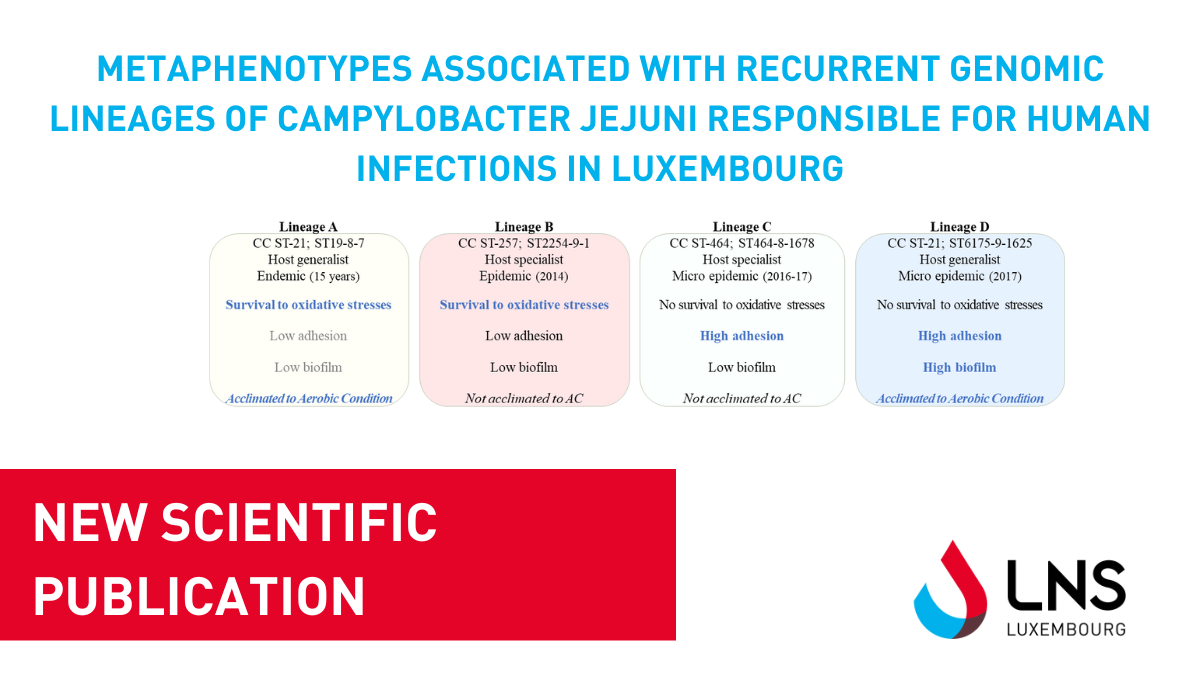
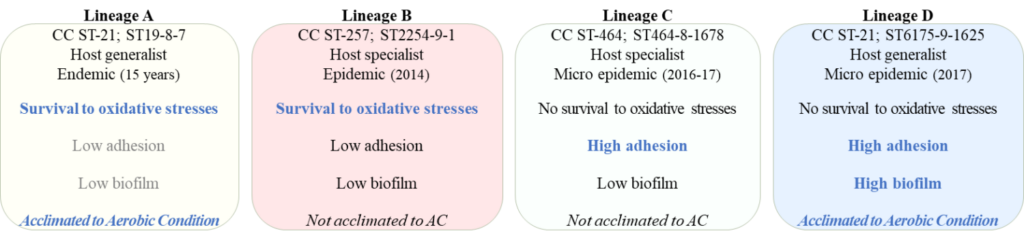
A new publication has been released exploring the behaviour of 4 Campylobacter jejuni genetic lineages commonly found in human infections and identified in Luxembourg as endemic.
Dr. Catherine Ragimbeau, Clinical Scientist in Bacterial Genomics for the Epidemiology and Microbial Genomics Unit, and the former PhD student Morgane Nenning, conducted this study with the objective of confirming the hypothesis that strains of the same lineage may exhibit adaptive behavior that favors spatio-temporal survival and persistence.
Find below the study the summary:
Campylobacter jejuni is the leading cause of bacterial diarrheal diseases around the globe but does not seem to be involved in large outbreaks like other foodborne pathogens. The species is characterized by a high degree of genetic diversity, mainly driven by horizontal genetic exchange. C. jejuni is a strict microaerobic, capnophilic, and thermotolerant pathogen. Given its growth requirements, its ability to persist in food environments and be transmitted throughout food processing has long puzzled scientists.
In our previous study, we have demonstrated that the genetic population structure of C. jejuni is partly composed of clonal expansion of some founders that persist over a long period spanning up to 13 years in Luxembourg. Considering the presence of these stable genotypes, we hypothesized that strains from the same lineage might share adaptive behaviors that could favor spatio-temporal survival and persistence. Using phenotypical assays in controlled conditions and functional genomic analyses from whole genome MLST data, we investigated the possible link between phenotypic traits and emergence or persistence of these genotypes. According to our analyses, a combination of independent biological responses was specific to each lineage. These data suggest that a combination of different phenotypical abilities, the metaphenotype, may contribute to the adaptation and survival of these monomorphic lineages over time and from various sources. These results also indicate the existence of a selective pressure behind the emergence and persistence of the most adapted and resistant lineages to different environmental stresses. From the functional genomic analyses conducted, the presence of a potentially functional T6 secretion system (SS) in the strains of lineage D might explain the propensity of these strains to be strong biofilm producers. Interestingly, one of the unique targets identified in this lineage was an ATPase called RloA, which potentially confers a completed T6SS machinery, while other lineages with T6SS genes lack such an ATPase.
The results suggested the existence of diffuse outbreaks caused by better-adapted and persistent C. jejuni strains to environmental stresses throughout the transmission route to human.
This study was part of the FNR-funded CampylOmic project (FNR-funded C17/BM/11684203) and was presented by Morgane Nennig for her thesis defence at LNS in March 2022.
Front Microbiol. Nennig M, Clément A, Longueval E, Bernardi T, Ragimbeau C, Tresse O. 2022 Sep 7;13:901192. doi: 10.3389/fmicb.2022.901192. PMID: 36160185; PMCID: PMC9490421.
Know more: